Abstract
In recent years, there has been an influx of new tracers into the field of nuclear medicine and molecular imaging. Most of those that have been Food and Drug Administration–approved for clinical imaging exploit various mechanisms of protein biochemistry and molecular biology to bring about their actions, such as amino acid metabolism, protein folding, receptor–ligand interactions, and surface transport mechanisms. In this review, we attempt to paint a clear picture of the basic biochemistry and molecular biology of protein structure, translation, transcription, posttranslational modifications, and protein targeting, in the context of the various radiopharmaceuticals currently used clinically, all in an easy-to-understand language for entry-level technologists in the field. Tracer characteristics, including indications, dosage, injection-to-imaging time, and the logic behind the normal and pathophysiologic biodistribution of these newer molecular tracers, are also discussed.
Proteins are the fundamental building blocks of every cell (1). They are made up of specific sequences of amino acids joined by peptide bonds and are arranged end to end in long chains called polypeptides. Two amino acids joined by a peptide bond is called a dipeptide, 3 amino acids linked by a peptide is a tripeptide, and so on, with an 8-amino-acid sequence making up an octapeptide as seen with the radiotracer 111In-octreotide (2). The sequence of amino acids in proteins is determined by the genetic code of the DNA. The gene sequence in the DNA is transcribed into the messenger RNA (mRNA) in the nucleus by an enzyme called RNA polymerase, with the help of a host of assisting enzymes called transcription factors. The initial mRNA contains sequences that code for the protein (exon) along with noncoding regions (introns), which are processed (spliced) to obtain the final mRNA with the correct sequence coding for the given protein. One gene may code for multiple proteins, whereby the same gene sequence is spliced out in a variety of different patterns to yield function proteins of differing sequences (alternative splicing). This process increases the diversity and the coding capacity of the genes. However, aberrant splicing reactions can result in disease conditions such as β-thalassemia, which is a severe blood disorder characterized by abnormal formation of hemoglobin (3).
Once spliced, the processed mRNA is then exported from the nucleus into the cytoplasm. On reaching the cytoplasm, the ribosomes (protein-producing molecular machines) hop onto the mRNA in search of a specific 3-nucleotide sequence called the start codon, which will act as a cue for the ribosome to start building the polypeptide chain on the basis of the subsequent nucleotide sequences. In eukaryotes (organisms with an intact nucleus, which includes everything from amoebas, worms, birds, and plants to humans), the start codon usually codes for the amino acid methionine (4). In prokaryotes (unicellular organisms without a nucleus, in which the DNA is floating in the cytoplasm, including members such as bacteria and archaea), it is a modified version of methionine (formyl methionine) (5). The nucleotide sequence in the mRNA is read in triplets (codon), and each codon codes for an amino acid. However, one amino acid may be called on by different codons with differing nucleotide sequences (degeneracy of the codon), and this property of the genetic code makes it more fault-tolerant for point mutations. As the ribosomes move down the mRNA, reading the codons, the amino acids are brought to the ribosomes by specific transfer RNAs that carry the corresponding amino acid and have matching anticodon nucleotide sequences that can correctly base-pair (form a covalent bond) to the codon on which the ribosome sits at any given moment. The new amino acid is then added to the methionine (or to formyl methionine) in a condensation reaction in which a molecule of water is removed to form a peptide bond (-CONH-) between the terminal carbon atom of the methionine (C-terminal) and the amino terminal of the next amino acid (N-terminal) (Fig. 1) (6).
Condensation reaction. Two amino acids are joined together to form peptide bond with release of water molecule.
The previous transfer RNA (which brought in the methionine) is released, and the ribosome along with the new transfer RNA now carrying the 2 amino acids (dipeptide) then proceeds to the next codon. The process is repeated to generate a tripeptide, tetrapeptide, pentapeptide, and so on, until a long polypeptide protein chain is created as prescribed by the genetic code. Once the ribosome reaches the stop codon with a sequence that does not code for an amino acid and no transfer RNAs are recruited, the ribosomes recruit a release factor enzyme that causes hydrolysis of the final C-terminal group of the polypeptide and attaches it to the ribosome, thus resulting in release of the full-length polypeptide chain. There are multiple ribosomes hurtling down the mRNA doing the translation simultaneously, one behind the other, resulting in many polypeptides being synthesized from an mRNA and thereby increasing the yield of the protein product manyfold. Once the protein chain is translated, it must then be folded in precise 3-dimensional conformations for it to become functionally active. This folding process is done cotranslationally by special molecular chaperone proteins that guide the nascent polypeptide chain to fold into its secondary, tertiary, and quaternary structures (7). The precise folding of the long polypeptide chain is important for correct forming of the protein’s active site where the catalyzing reaction occurs or for stable incorporation of the necessary ion or chemical group (cofactor) to achieve its designated biologic task (8). The secondary structure of the protein is formed by the meticulous folding of the peptide chain into a helix or a pleated sheet. This process is mediated by the specific ϕ- and ψ-torsion angles of the amino acids that would result in hydrogen bonding of the adjacent groups of the amino acids in the vicinity. The order of the amino acids specified by the genetic code dictates this folding process, which would result in an energetically favorable (less entropy) stable conformation (Fig. 2) (9).
Protein secondary structure formation. Precise folding of polypeptide chain is achieved by rotational angles (φ, ψ) of backbone bonds flanking central α-carbon atom of each amino acid. These rotational angles are specific for each amino acid and are instrumental in shaping protein structure as prescribed by genetic code.
The chain of helices and sheets is further folded in 3-dimensional space and stabilized by hydrogen bonds and ionic interactions between atoms within the chain and within the watery (aqueous) environment in the cytoplasm (Fig. 3).
Protein tertiary structure: intra- and intermolecular bonds help form and stabilize precise 3-dimensional protein structure into helices and sheets.
Once the chain is correctly folded into its tertiary structure, it may then need to join cooperatively with one or more folded peptide chains to form the final functional quaternary structure (e.g., hemoglobin is made up of 4 folded polypeptide subunits with an iron group stabilized in the middle of each subunit) (Fig. 4) (10,11). Similarly, an antibody protein, such as the one used in the non-Hodgkin lymphoma radioimmunotherapy agent 90Y-ibritumomab tiuxetan, is made up of 4 different polypeptide chains (primary structure) that are folded into β-barrels (secondary structures). This folding in turn makes it possible for those from the same and adjacent chains to form bonds with each other called disulfide bridges (tertiary structure), thus giving the antibody molecule its final 3-dimensional Y shape (quaternary structure) (12).
Protein quaternary structure: helices and pleated sheets of different polypeptide chains may further associate together to form quaternary structures as in case of hemoglobin molecule in red blood cells. Each hemoglobin molecule is composed of 4 polypeptide subunits (2 α-chains and 2 β-chains), each stabilized by ion group (heme) in center.
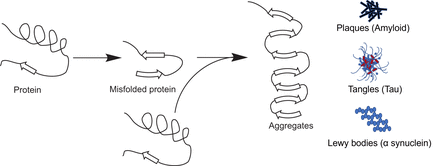
If the protein is not correctly folded, it may be destroyed by specialized enzymes called proteasomes (13). In some pathologic situations, this misfolding of proteins may result in aberrant protein aggregates such as those seen in Alzheimer disease, where the proteins amyloid β and tau are incorrectly folded, resulting in sticky tarlike plaques in the neuronal tissue of the brain and further leading to inflammation and associated pathology (14). These misfolded protein aggregates in the affected neurons of the brain are the targets of the Alzheimer detection agents 18F-florbetaben, 18F-florbetapir, and 18F-flutemetamol, all of which detect amyloid β-plaques (15), and 18F-flortaucipir, which detects misfolded tau protein tangles (Fig. 5) (16).
Protein misfolding can lead to pathology. Correct folding of protein into its proper 3-dimensional structure is important to function correctly. Incorrectly folded proteins either are destroyed by proteasomes or may form insoluble aggregates such as plaques, tangles, and Lewy bodies, which can lead to pathologic conditions as in Alzheimer disease.
Once the protein is correctly folded, it either stays in the cytoplasm or is exported outside the cell to its correct destination in the body. This process is guided by the types of amino acid residues, called signal sequences, in the N-terminal region of the polypeptide chain (e.g., hormones such as insulin are produced by the pancreas and are secreted into the blood for blood sugar regulation, whereas digestive enzymes are secreted by stomach cells into the gut for the task of digestion) (17).
Some proteins are shunted to the cell surface to be part of the cell membrane to act as switches (also known as receptors) for transmitting a signal into the cell’s nucleus. The signal starts a specific cellular function based on environmental cues or based on another specific outside protein, such as a hormone or neurotransmitter, binding to it (18). These receptors have extremely specialized functions, such as when cluster-of-differentiation (CD) receptor protein CD20 (detected by the tracer 90Y-ibritumomab tiuxetan) on the surface of B lymphocytes helps to produce antibodies (19), or when the CD206 protein (bound by the lymphoscintigraphy mannose sugar tracer 99mTc-tilmanocept) on the surface of macrophages scavenges sugar molecules from pathogens (20). Sometimes a receptor is synthesized in many almost-identical forms (also known as subtypes) to achieve a variety of functions in different organs by the same activator ligand, as is the case with several subtypes of somatostatin receptor (SSTR): SSTR1, SSTR2 (bound by the neuroendocrine tumor theranostic agents 68Ga/177Lu/64Cu-DOTATATE, 68Ga/177Lu/64Cu-DOTATOC, and 111In-octreotide), SSTR3 (bound by 111In-octreotide), SSTR4, and SSTR5 (bound by 68Ga/177Lu/64Cu-DOTATATE, 68Ga/177Lu/64Cu-DOTATOC, and 111In-octreotide), all of which, in response to somatostatin, inhibit a variety of cell growth and other activities from their cell surface locations depending on the organ (or cancer) in which they are present (21). Such versatile protein receptors are sometimes localized to the cytoplasm itself, such as the estrogen receptors (bound by the breast carcinoma estrogenlike synthetic tracer 18F-fluoroestradiol), which would need the activating estradiol molecule (serving as a ligand) to cross the cell membrane and bind to the receptors. This causes the receptors to physically move into the nucleus and start the transcription of a multitude of genes regulating many important body functions, such as cell proliferation and bone health (22).
Not all surface proteins take up the role of being receptors for cell-signaling activities. Some function as carriers of other molecules across the cell membrane, such as when the dopamine active transporter helps to transport (reuptake) the secreted excitatory neurotransmitter dopamine back into the neuron (23). Also taken up into intact neurons in the brain is the Parkinson disease tracer 123I-ioflupane, which is analogous to dopamine (24). Similarly, the uptake-1 transporter helps the reuptake of the neurotransmitter norepinephrine back into the neuron. This reaction also occurs with the neuroendocrine tumor theranostic agent 123/131I-meta-iodobenzylguanidine (MIBG), which is analogous to norepinephrine (25,26).
Cell surface protein transporters act as transporters of not only neurotransmitters but also a variety of other biomolecules, including amino acids, which are needed for protein synthesis. In humans, there are 10 different types of amino acid transporters. Some of these, including L-type amino acid transporter and alanine-serine-cysteine transporter 2, in increased numbers are seen transporting large amounts of amino acids, such as leucine, into the cells for increased protein synthesis due to high demand in a cancerous state (27). A similar reaction occurs with the artificial nonmetabolizable amino acid leucin analog tracer 18F-fluciclovine for prostate cancer imaging (28).
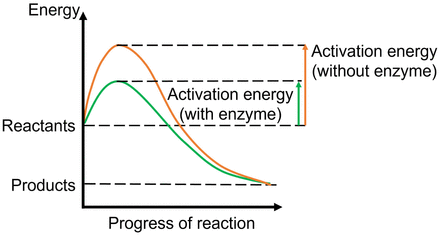
Some proteins, on the other hand, act as enzyme catalysts. A protein enzyme catalyzes a reaction by creating a conducive and protective environment in its active site that will produce an ideal condition for the chemical reaction to proceed (decrease of activation energy for starting the chemical reaction, resulting in the product’s becoming more stable than the reactants) (Fig. 6) (29). These principles govern all the chemical reactions that take place in our body for the reactions to work in a complex chemical environment. An example of a cell surface transport protein that also acts as an enzyme is the prostate-specific membrane antigen (PSMA) seen on normal prostate cells, as well as on some other organs, including the kidneys, small intestine, and nervous system. PSMA enzymatically acts on dietary folic acid (vitamin B9) and on the neurotransmitter N-acetylaspartylglutamic acid to releases the amino acid glutamate, which helps mobilize calcium to support normal cell growth in the prostate and maintain neuronal functions in the brain (30,31). The increased presence of PSMA on prostate cancer cells is the target for the imaging agents 68Ga-PSMA11 and 18F-piflufolastat, which were approved by the Food and Drug Administration in 2020 and 2021, respectively.
Enzyme as catalysts. Enzymes catalyze chemical reactions by lowering activation energy required for reactants to progress through steps of chemical reaction. This lowering of energy includes that of the high-energy transition state at the peak of the energy profile, which is lower when enzymes are present, hence making it easier for reaction to progress.
MOLECULAR TRACER USING AMINO ACID METABOLISM (18F-FLUCICLOVINE (AXUMIN; BLUE EARTH DIAGNOSTICS) (32))
18F-fluciclovine is a synthetic amino acid that resembles leucine and is labeled with the radionuclide 18F. It was approved by the Food and Drug Administration in 2016 for imaging of prostate cancer recurrence with increased blood levels of prostate-specific antigen. 18F-fluciclovine is carried across the cancer cell’s membrane by the L-type amino acid transporter and by alanine-serine-cysteine transporter 2, which are seen in higher amounts in cancer cells. The driving force for this tracer uptake, apart from the increased need of the cancer cells for amino acids for protein building, are the androgens (male sex hormone produced in the testicles), which in general are dangerous proponents of cancer growth. In one type of treatment for prostate cancer, biochemical castration, drugs are given that shut down production of this hormone in the male gonads. However, nature may eventually bypass this effect in some patients by producing androgen from sources outside the testicles or through DNA-changing mutations, eventually resulting in recurrence of prostate cancer and metastasis. This condition is clinically called metastatic castration-resistant prostate cancer. 18F-fluciclovine, once transported into the prostate cancer cells, gets trapped in the cells. Being an artificial amino acid, it cannot be used for protein building by the cell’s transfer RNAs and ribosomes (Table 1).
Clinical Properties of Current Food and Drug Administration–Approved Molecular Tracer (18F-Fluciclovine) Using Amino Acid Metabolism
MOLECULAR TRACERS DETECTING PROTEIN FOLDING OR MISFOLDING
18F-Florbetapir (Amyvid; Lilly), 18F-Florbetaben (Neuraceq; Life Molecular Imaging), and 18F-Flutemetamol (Vizamyl; GE Healthcare) (33–35)
18F-florbetapir, 18F-florbetaben, and 18F-flutemetamol bind to the misfolded β-amyloid proteins forming plaques in the brain of Alzheimer disease patients, specifically plaques that appear in the gray matter of the outer cerebral cortex, where β-protein should not be present (Table 2).
Clinical Properties of Current Food and Drug Administration–Approved Molecular Tracers Detecting Protein Folding or Misfolding
MOLECULAR TRACERS TARGETING CELL SURFACE PROTEIN RECEPTORS
18F-Fluoroestradiol (Cerianna; Zionexa) (37)
18F-fluoroestradiol is indicated for use in PET imaging for the detection of estrogen receptor–positive lesions in patients with recurrent or metastatic breast cancer. 18F-fluoroestradiol is a synthetic estrogen analog that migrates to the estrogen receptor proteins in the cytoplasm of breast cancer cells (Table 3).
Clinical Properties of Current Food and Drug Administration–Approved Molecular Tracers Targeting Cell Surface Protein Receptors
99mTc-Tilmanocept (Lymphoseek; Cardinal Health) (38)
99mTc-tilmanocept is indicated for use with or without scintigraphic imaging for lymphatic mapping using a handheld γ-probe to locate lymph nodes draining a primary solid-tumor site as a component of intraoperative management. It is also indicated with or without scintigraphic imaging for guiding a sentinel lymph node biopsy using a handheld γ-probe in patients with clinically node-negative squamous cell carcinoma of the oral cavity or with breast cancer or melanoma. 99mTc-tilmanocept is made up of many units of diethylenetriamine pentaacetic acid (kidney-imaging agent) and mannose (a type of sugar), linked together on a carbohydrate dextran backbone to form a giant molecule that is able to bind to the CD20 receptor protein on the surface of macrophages (a type of disease-fighting blood cell constantly patrolling the blood and lymph nodes). When injected under the skin, 99mTc-tilmanocept drains into the lymph and accumulates in the macrophages in the sentinel lymph node and beyond (Table 3).
68Ga/177Lu/64Cu-DOTATATE (NETSPOT [Advanced Accelerator Applications]/LUTATHERA [Advanced Accelerator Applications]/Detectnet [Curium]), 68Ga/177Lu/64Cu-DOTATOC, 68Ga/177Lu/64Cu-DOTANOC, and 111In-Octreotide (OctreoScan; Curium) (39–42)
The molecular agents 68Ga/177Lu/64Cu-DOTATATE, 68Ga/177Lu/64Cu-DOTATOC, 68Ga/177Lu/64Cu-DOTANOC, and 111In-octreotide bind to protein receptors on the surface of SSTR-containing neuroendocrine tumors. This binding is selective for the subtype of SSTR, with radiolabeled DOTATATE and DOTATOC binding to SSTR2 and SSTR5 whereas 111In-octreotide binds to all 3 SSTR subtypes (SSTR2, SSTR3, and SSTR5). The radiolabeled DOTATATE fits the classic definition of a theranostic radiopharmaceutical, in that the same agent can be used for both diagnostic (68Ga-DOTATATE) and therapeutic (177Lu/64Cu-DOTATATE) purposes (Table 3).
90Y-Ibritumomab Tiuxetan (Zevalin; Acrotech Biopharma) (43)
The antibody radiopharmaceutical 90Y-ibritumomab tiuxetan binds to CD20 receptor protein molecules on mature and malignant B lymphocytes, a type of disease-fighting cell in the blood, while sparing the immature and parent cells in the bone marrow. Although the function of the CD20 receptor on B cells is unknown, it is thought to play a role in calcium entry across the B-cell membrane and in maintenance of the required amounts of calcium inside the cell for activation in the disease-fighting process, including antibody production. The big advantage of anti-CD20 antibodies is that they attack only malignant B cells and not the precursor cells, thus preventing the patient from losing all B cells, avoiding even more negative outcomes. The β-radiation from the 90Y isotope then destroys the cancerous B cells (Table 3).
MOLECULAR IMAGING TRACERS TARGETING CELL SURFACE PROTEIN TRANSPORTERS
123I-Ioflupane (DaTscan; GE Healthcare) (44)
123I-ioflupane binds to presynaptic dopamine transporters, seen in the striatal region of the brain. A defining feature of Parkinson disease is a marked reduction in the dopamine-secreting neurons in this portion of the brain. Dopamine, once secreted into the synaptic gap between the presynaptic and postsynaptic neurons (neurons sending and receiving the nerve signals, respectively), is reabsorbed back into the presynaptic neuron through the dopamine transporter proteins on the presynaptic neuron endings, thus preventing continuous excitation. The nerve signals are related to higher-order functions of the brain, including movement and coordination. In the absence or destruction of these neurons, movement disorders typical to those of Parkinson syndromes will result. By using a tracer that binds to the presynaptic dopamine transporters, one can obtain a quantitative measure and spatial distribution of these transporters and, hence, the dopamine secreting neurons (Table 4).
Clinical Properties of Current Food and Drug Administration–Approved Molecular Tracers Targeting Cell Surface Protein Transporters
123/131I-Iobenguane (AdreView [GE Healthcare]/Azedra; Progenics Pharmaceuticals) (45)
123/131I-iobenguane, also known as MIBG, is structurally similar to the fight-or-flight hormone norepinephrine. Once secreted into the synapse between 2 neurons to facilitate neurotransmission, norepinephrine is taken back up into the presynaptic neurons through transporters called uptake 1. Uptake 1 is normally found in tissues but is overexpressed in certain neuroendocrine tumors, such as pheochromocytomas and neuroblastomas. Since MIBG resembles norepinephrine, it will be taken up in neuroendocrine tumor cells along with the neurotransporter in a higher amount. The radiolabeled MIBG is also a theranostic radiopharmaceutical; the same agent can be used for both diagnostic (123I-MIBG) and therapeutic (131I-MIBG) purposes (Table 4).
68Ga-PSMA11 and 18F-Piflufolastat (Pylarify; Lantheus) (46,47)
68Ga-PSMA11 and 18F-piflufolastat attach to the outer portion (extracellular domain) of the PSMA found in elevated levels (100- to 1,000-fold) on the surface of 95% of prostate cancer cells (48). PSMA is also normally seen at low levels in the kidneys, liver, tear glands, salivary glands, spleen, and nervous tissue. Once the radiotracer binds to the surface membrane protein, PSMA—along with the attached radiotracer—is transported into the cell, thus trapping the tracer within the cancer cell (Table 4) (49).
CONCLUSION
If past decades in nuclear medicine saw bold moves in physics and instrumentation, the future landscape is evolving to be the scintillating age of molecular and systems biology. The present and future growth of molecular imaging and therapy is based on using the intricate biologic and biochemical pathways of the human body. For the nuclear medicine technologist to appreciate the interactions of radiopharmaceuticals within the body and thus delineate the best information possible, a good understanding of the underlying mechanisms is required. This knowledge should be grounded in sound fundamentals of molecular processes in living systems, which will enable the technologist to integrate theory and processes into the realm of applied clinical practice. This ability will also engender a new generation of professionals who eagerly seek out cutting-edge knowledge and challenges that may elude the average health-care professional, leading the profession to the 21st century.
DISCLOSURE
No potential conflict of interest relevant to this article was reported.
Footnotes
CE credit: For CE credit, you can access the test for this article, as well as additional JNMT CE tests, online at https://www.snmmilearningcenter.org. Complete the test online no later than September 2025. Your online test will be scored immediately. You may make 3 attempts to pass the test and must answer 75% of the questions correctly to receive Continuing Education Hour (CEH) credit. Credit amounts can be found in the SNMMI Learning Center Activity. SNMMI members will have their CEH credit added to their VOICE transcript automatically; nonmembers will be able to print out a CE certificate upon successfully completing the test. The online test is free to SNMMI members; nonmembers must pay $15.00 by credit card when logging onto the website to take the test.
Published online Feb. 23, 2022.
REFERENCES
- Received for publication January 11, 2022.
- Accepted for publication February 3, 2022.