Abstract
Radiopharmaceutical dosimetry is usually estimated via organ-level MIRD schema-style formalisms, which form the computational basis for commonly used clinical and research dosimetry software. Recently, MIRDcalc internal dosimetry software was developed to provide a freely available organ-level dosimetry solution that incorporates up-to-date models of human anatomy, addresses uncertainty in radiopharmaceutical biokinetics and patient organ masses, and offers a 1-screen user interface as well as quality assurance tools. The present work describes the validation of MIRDcalc and, secondarily, provides a compendium of radiopharmaceutical dose coefficients obtained with MIRDcalc. Biokinetic data for about 70 currently and historically used radiopharmaceuticals were obtained from the International Commission on Radiological Protection (ICRP) publication 128 radiopharmaceutical data compendium. Absorbed dose and effective dose coefficients were derived from the biokinetic datasets using MIRDcalc, IDAC-Dose, and OLINDA software. The dose coefficients obtained with MIRDcalc were systematically compared against the other software-derived dose coefficients and those originally presented in ICRP publication 128. Dose coefficients computed with MIRDcalc and IDAC-Dose showed excellent overall agreement. The dose coefficients derived from other software and the dose coefficients promulgated in ICRP publication 128 both were in reasonable agreement with the dose coefficients computed with MIRDcalc. Future work should expand the scope of the validation to include personalized dosimetry calculations.
Accurate estimations of absorbed radiation dose are required to ensure patient safety in nuclear medicine. The MIRD schema (1) provides a computational basis for assessing absorbed dose at the organ level, tissue level, cellular level, or various spatial subdivisions; however, organ-level dosimetry remains the prevailing methodologic paradigm for routine clinical dose assessment.
Implementation of the MIRD schema or equivalent formalisms in dosimetry software generally requires 2 underlying elements: a mathematic representation of anatomy known as a computational phantom, and a database of radionuclide S values (1) specific to the phantom (usually derived via Monte Carlo simulation). Reference phantoms, which model representative individuals of particular sex and age groups, have undergone several important revisions over the past 50 y. First, the consensus anatomic reference data have been revised, with the data of the International Commission on Radiological Protection (ICRP) publication 23 (2) being superseded by ICRP publication 89 (3). Second, the computerized formats for depicting anatomy have been refined.
Recently, the MIRDsoft project was initiated to provide the community with a suite of free dosimetry software programs endorsed by the MIRD committee of the Society of Nuclear Medicine and Molecular Imaging. MIRDcalc, the flagship program in the MIRDsoft suite, is a Microsoft Excel–based executable program implementing the MIRD schema at the organ level. It incorporates up-to-date anatomic models, including the ICRP publication 110 series adult computational reference phantoms (4) and the ICRP publication 143 series pediatric reference phantoms (5,6). These phantoms offer numerous advantages over the first-generation stylized phantoms, including anatomically realistic organ shapes and interorgan spacing. MIRDcalc also factors uncertainty into the dose coefficients by propagating uncertainties in the input biokinetics and organ masses. Additional features include a single-screen interface, quality assurance tools, and batch processing capabilities.
An overview of the development and features of MIRDcalc is provided in part 1 of this 2-part article (7). In this second part, the primary aim was to comprehensively benchmark the dose coefficients obtained from MIRDcalc against a compendium of published reference dose coefficients (8) and the output of other reference dosimetry software.
COMPUTATION OF DOSIMETRIC QUANTITIES
Absorbed Dose
The present investigation considers organ-level mean absorbed doses and assumes that the masses of phantom regions remain constant over the period of irradiation. The organ-level time-independent formulation of the MIRD schema therefore applies (1). Given a target region  irradiated over a time period
irradiated over a time period  , the absorbed dose coefficient
, the absorbed dose coefficient  (mGy/MBq) is defined as the absorbed dose (mGy) normalized to the administered activity (MBq):
(mGy/MBq) is defined as the absorbed dose (mGy) normalized to the administered activity (MBq): Eq. 1Here,
Eq. 1Here,  is the time-integrated activity coefficient (TIAC), and
is the time-integrated activity coefficient (TIAC), and  is known as the S value (or S coefficient), the absorbed dose rate to
is known as the S value (or S coefficient), the absorbed dose rate to  per unit activity in source region
per unit activity in source region 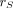 . S values are specific to a radionuclide and computational phantom. For monoenergetic emissions:
. S values are specific to a radionuclide and computational phantom. For monoenergetic emissions: Eq. 2where
Eq. 2where  is the energy of the ith nuclear transition,
is the energy of the ith nuclear transition,  is the number of ith transitions per nuclear transformation, and
is the number of ith transitions per nuclear transformation, and  is the specific absorbed fraction (SAF) (kg−1). The SAF represents the fraction of
is the specific absorbed fraction (SAF) (kg−1). The SAF represents the fraction of  absorbed in
absorbed in  for transition i occurring in source
for transition i occurring in source  , normalized to the mass of the target region. For β-emissions, the spectrum of energies is considered:
, normalized to the mass of the target region. For β-emissions, the spectrum of energies is considered: Eq. 3where
Eq. 3where  is the β-endpoint energy, and
is the β-endpoint energy, and  is the distribution for the number of β-particles emitted per megaelectron volt per nuclear transformation, as a function of energy.
is the distribution for the number of β-particles emitted per megaelectron volt per nuclear transformation, as a function of energy.
The TIAC represents the number of nuclear transformations occurring in a source region over a specified time period  :
: Eq. 4where
Eq. 4where  is the time-dependent activity in
is the time-dependent activity in  , and
, and  is the administered activity.
is the administered activity.
All programs used in this work use the MIRD or mathematically equivalent formalism for absorbed dose computation.
Effective Dose
The effective dose (mSv) is considered a measure of risk related to radiation-induced stochastic effects. It is a sex-averaged, tissue-weighted sum of mean organ equivalent doses (9). The effective dose coefficient (mSv/MBq) (i.e., effective dose per unit administered activity) can be calculated as… Eq. 5where
Eq. 5where  is the tissue-weighting factor for tissue T, and
is the tissue-weighting factor for tissue T, and  is the equivalent dose, wherein the radiation-weighting factor
is the equivalent dose, wherein the radiation-weighting factor  accounts for the differential biologic effectiveness of each radiation type R:
accounts for the differential biologic effectiveness of each radiation type R: Eq. 6
Eq. 6
DOSIMETRY SOFTWARE AND PHANTOMS
MIRDcalc
MIRDcalc is Excel-based software freely obtainable at www.mirdsoft.org. For dose computation, MIRDcalc uses a database of S values for the ICRP publication 110 (4) reference adult phantoms and ICRP publication 143 (6) reference pediatric phantoms (Fig. 1). The reference phantom S-value database was generated using SAFs promulgated by the ICRP (10,11). The SAFs were used in combination with the radionuclide decay data of ICRP publication 107 (12) to compute the S values (Eq. 2); in the case of β-particles, the full spectrum was used (Eq. 3). Effective dose coefficients are computed using the tissue-weighting factors of ICRP publication 103 (13).
Phantoms used to derive dose coefficients compared in this work. Solid arrows denote comparisons using different phantoms and different software. Dashed arrows represent comparisons using same phantoms but different software. DC = dose coefficient; ORNL = Oak Ridge National Laboratory; wT = tissue-weighting factor.
In addition to the S values for each phantom, the organ masses and organ fractional blood content are stored within MIRDcalc for use in various computations. The organ masses and fractional blood content of the adult phantoms are defined in ICRP publication 89. For the pediatric phantoms, the suggested reference values of Wayson et al. (14) were used, which, notably, account for volumetric changes and changes in blood vascularization across the different reference ages. A summary of all phantom organ, parenchyma, and blood masses used in MIRDcalc is presented as supplemental material in part 1 of this article (supplemental materials are available at http://jnm.snmjournals.org) (7).
IDAC-Dose
IDAC-Dose 2.1 (15) organ-level dosimetry software is MATLAB-based and uses S values for the ICRP publication 110 adult reference phantoms (Fig. 1) for dose computation; similar to MIRDcalc, IDAC S values were derived from ICRP publication 133 SAFs. Effective dose coefficients are computed using ICRP publication 103 tissue-weighting factors. IDAC-Dose is freely obtainable from www.idac-dose.org.
Notably, IDAC-Dose and MIRDcalc are based on identical phantoms.
OLINDA
OLINDA 2.1, preceded by OLINDA 1.0 and MIRDOSE software, is Java-based. It uses an S-value database derived from the Radiation Dose Assessment Resource (RADAR) phantom series (Fig. 1) (16,17). Effective dose coefficients are computed using ICRP publication 103 tissue-weighting factors. OLINDA is obtainable under license from Hermes Medical Solutions (www.hermesmedical.com). Only the RADAR reference adult male and female phantoms were used for comparisons in this work.
The RADAR phantoms of OLINDA and the ICRP publication 110 phantoms of MIRDcalc are based on the same reference organ parenchyma masses but possess different organ shapes and positioning. The RADAR phantoms do not contain a blood source region or account for organ fractional blood content. For walled organs, a different dose estimation methodology is used (18,19).
REFERENCE DOSE COEFFICIENTS AND BIOKINETIC DATA
ICRP publication 128 is a compendium of reference dosimetric information for about 70 currently or historically used radiopharmaceuticals. For certain radiopharmaceuticals, several use scenarios or administration routes are considered. Organ-level TIACs and mean organ-absorbed dose coefficients are provided for the Cristy–Eckerman stylized mathematic reference adult and pediatric phantoms (15-, 10-, 5-, and 1-y-old) (20). The effective dose coefficients reported in ICRP publication 128 were computed using ICRP publication 60 tissue-weighting factors.
The Cristy–Eckerman phantoms used to compute the dose coefficients reported in ICRP publication 128 possess similar organ masses to the phantoms of MIRDcalc but have drastically different shapes and different interorgan spacing. For walled organs, a different dose estimation methodology is used (18,19).
MODIFICATIONS OF ICRP PUBLICATION 128 REFERENCE BIOKINETIC DATA FOR USE IN OTHER PHANTOMS
The ICRP publication 128 TIACs are the biokinetic source data used in all our calculations. However, these TIACs are specific to the source regions of the Cristy-Eckerman stylized reference phantoms; some of these source regions have been redefined or modified in more modern reference phantoms, such as those used in MIRDcalc, IDAC, or OLINDA. Therefore, TIACs for the Cristy–Eckerman phantom source regions were transposed into the other phantom source regions using the approach of Andersson et al. (21). Specific assumptions and adjustments are detailed here.
Colon Wall
The colon wall of the Cristy-Eckerman phantoms is partitioned into 2 regions: the upper large intestine, comprising the ascending and transverse colon, and the lower large intestine, comprising the descending and sigmoid colon. In modern reference phantoms, the colon wall is instead partitioned into 3 regions consonant with the updated ICRP human alimentary tract model: the right colon, comprising the ascending colon and the proximal half of the transverse colon; the left colon, comprising the descending colon and the distal half of the transverse colon; and the rectosigmoid colon. TIACs provided in ICRP publication 128 for the walls of the upper or lower large intestine were transposed into the modern phantoms using the following formulas (20): Eq. 7
Eq. 7 Eq. 8
Eq. 8 Eq. 9
Eq. 9
Colon Contents
The colon content source regions follow the same partitioning scheme as above, but with different coefficients required to account for the region masses: Eq. 10
Eq. 10 Eq. 11
Eq. 11 Eq. 12
Eq. 12
Walled Organs (General Case)
In MIRDcalc and IDAC-Dose, which have the option to assign TIACs to the entire wall or constituent subregions, the entire wall was considered as the source.
OLINDA does not support the use of walled organs as source regions; only the contents of walled organs may be used as sources. When ICRP publication 128 provided a TIAC for a wall source, this was appended to the TIAC for the other-organs-and-tissues region. We note that this approach may critically underestimate the absorbed dose coefficients for radiopharmaceuticals with elevated uptake or prolonged retention in walled organs.
Skeleton
For radiopharmaceuticals for which ICRP publication 128 explicitly specified TIACs for the cortical or trabecular bone source regions, those values were used without modification. When a TIAC was specified for a generic bone-surface source region (i.e., bone surface–seeking radionuclides), the TIAC was reapportioned to the cortical and trabecular bone surfaces in a 40:60 ratio for the adult, 15-y-old, or 10-y-old phantoms or in a 30:70 ratio for the 5-y-old or 1-y-old phantoms (8). For bone volume–seeking radionuclides, 80:20 and 60:40 cortical-to-trabecular ratios, respectively, were used to define bone volume sources for the age groups above (8).
Blood
A salient difference among the dosimetry programs used here relates to the treatment of blood. The stylized phantoms used in the ICRP publication 128 dose calculations did not directly define a blood source; instead, ignoring differences in fractional blood content among organs, a blood source was approximated using a uniform whole-body source region for those calculations. Similarly, the hybrid phantoms of OLINDA do not directly implement a blood source. In IDAC-Dose, a blood source is defined and is based on ICRP publication 133 SAFs for total-body blood content; this blood source was designed to be used in combination with organ TIACs supplied for the organ parenchyma only (i.e., exclusive of blood contained within the organs). The ICRP publication 133 blood source region is available in MIRDcalc. MIRDcalc also contains novel dynamic blood and parenchyma regions to support either blood-inclusive or blood-exclusive TIAC inputs. Namely, this feature supports 2 areas of dosimetry research: the first is routine nuclear medicine dosimetry, with blood-inclusive organ uptake being quantified via imaging (e.g., volume-of-interest delineation, with the volume including organ parenchyma and organ blood content), and the second is radiation protection, with TIACs for organ parenchyma often being obtained through pharmacokinetic modeling. The MIRDcalc dynamic blood model actively removes blood and parenchyma subregions from the dynamic rest-of-blood and rest-of-parenchyma source regions, respectively, as well as from the rest-of-body source region, as individual organ TIACs are entered. To implement the dynamic blood model in MIRDcalc, SAFs for blood regions including heart contents, major vessels, and miscellaneous connective tissues were required. These SAFs were computed via methods described previously (10).
ICRP publication 128 considers 2 separate approaches for specifying TIACs associated with the blood—either explicit or implicit assignment. For radiopharmaceuticals for which a blood source is not explicitly given in ICRP publication 128, the ICRP publication 128 organ TIACs are assumed to apply to the whole organ (i.e., both parenchyma and blood contained within the organ). In contrast, for radiopharmaceuticals for which a blood source is explicitly given by the pharmacokinetic model, the organ TIACs are assumed to apply to the parenchyma only. Specific methods used for blood source input in each type of software are described in this article.
In IDAC-Dose, the blood, organ, and other-organs-and-tissues TIACs for each ICRP publication 128 radiopharmaceutical were input directly.
In OLINDA, when a blood source TIAC was available for a radiopharmaceutical, the total body was used as a surrogate for the blood. This was accomplished by running 2 separate calculations: the first was a calculation in which only the blood TIAC was input into the whole-body region (i.e., approximating the blood distribution as uniform throughout the body), and the second was a calculation in which the organ and ICRP publication 128 other-organs-and-tissues TIACs were input. The 2 respective dose calculations were then summed. For radiopharmaceuticals for which no explicit blood TIAC was given, a single calculation was run normally.
In MIRDcalc, when ICRP publication 128 provided a blood source TIAC, this was assigned to the classic ICRP blood source, and the ICRP publication 128 other-organs-and-tissues TIAC was assigned to the rest-of-parenchyma source region to avoid double-counting (i.e., to avoid source region overlap). For radiopharmaceuticals in the absence of a blood source, the ICRP publication 128 other-organs-and-tissues TIAC was assigned to the rest-of-body source region. In the latter case, the rest-of-body TIAC is distributed to its constituent regions in proportion to the blood-inclusive region masses.
COMPARISON OF DOSE COEFFICIENTS
Relative differences in dose coefficients are presented on the basis of 2 metrics. First, we define the logarithmic relative difference metric: Eq. 13where, for radiopharmaceutical
Eq. 13where, for radiopharmaceutical  ,
,  is the log relative difference (we refer to these here as Δ-values),
is the log relative difference (we refer to these here as Δ-values),  is the dose coefficient for target organ
is the dose coefficient for target organ  computed by MIRDcalc, and
computed by MIRDcalc, and  is the corresponding dose coefficient computed by another software/data source. The approach of quantifying differences via the log relative difference was selected to alleviate dependence on reference choice; the magnitude of the reported log relative differences is equivalent regardless of which method is chosen as the reference. Second, we consider the traditional percentage error, with MIRDcalc taken as the gold standard reference:
is the corresponding dose coefficient computed by another software/data source. The approach of quantifying differences via the log relative difference was selected to alleviate dependence on reference choice; the magnitude of the reported log relative differences is equivalent regardless of which method is chosen as the reference. Second, we consider the traditional percentage error, with MIRDcalc taken as the gold standard reference: Eq. 14We note that in the second case, the relative error approaches the log relative difference for very small differences, namely,
Eq. 14We note that in the second case, the relative error approaches the log relative difference for very small differences, namely, Eq. 15 Dose coefficients for 116 radiopharmaceutical-use scenarios were obtained with MIRDcalc dosimetry software and compared against the established reference data of ICRP publication 128 and against the output of other validated software (IDAC-Dose and OLINDA). Of the radiopharmaceuticals considered in ICRP publication 128, we have selected the subset with U.S. Food and Drug Administration approval as of the year 2020 for presentation in most of the figures and tables in the print version of this article (30 radiopharmaceuticals). Effective dose coefficients and associated relative differences are provided in Table 1 for adult reference phantoms; for pediatric phantoms, the corresponding data are given in Supplemental Tables 1–4. Graphical presentation (heat maps) of log relative differences in organ-absorbed dose coefficients are provided for reference adults in Figures 2 and 3. Corresponding heat maps for pediatric reference phantoms are given in Supplemental Figures 1–4. The underlying organ-absorbed dose coefficients for all phantoms and all ICRP publication 128 radiopharmaceuticals are available in the Supplemental Dose Coefficient Compendium.
Eq. 15 Dose coefficients for 116 radiopharmaceutical-use scenarios were obtained with MIRDcalc dosimetry software and compared against the established reference data of ICRP publication 128 and against the output of other validated software (IDAC-Dose and OLINDA). Of the radiopharmaceuticals considered in ICRP publication 128, we have selected the subset with U.S. Food and Drug Administration approval as of the year 2020 for presentation in most of the figures and tables in the print version of this article (30 radiopharmaceuticals). Effective dose coefficients and associated relative differences are provided in Table 1 for adult reference phantoms; for pediatric phantoms, the corresponding data are given in Supplemental Tables 1–4. Graphical presentation (heat maps) of log relative differences in organ-absorbed dose coefficients are provided for reference adults in Figures 2 and 3. Corresponding heat maps for pediatric reference phantoms are given in Supplemental Figures 1–4. The underlying organ-absorbed dose coefficients for all phantoms and all ICRP publication 128 radiopharmaceuticals are available in the Supplemental Dose Coefficient Compendium.
Adult Effective Dose Coefficients and Relative Differences Compared Among MIRDcalc, IDAC-Dose, and ICRP Publication 128
Organ-level absorbed dose coefficients for adult male compared via log relative differences (Eq. 13). (Top) ICRP publication 128 compared against MIRDcalc. (Middle) IDAC-Dose compared against MIRDcalc. (Bottom) OLINDA 2.1 compared against MIRDcalc. Red indicates a dose coefficient estimate higher than that of MIRDcalc; blue indicates lower. Black indicates off-scale values ( > 300).
> 300).
Organ-level absorbed dose coefficients for adult female compared via log relative differences (Eq. 13). (Top) ICRP publication 128 compared against MIRDcalc. (Middle) IDAC-Dose compared against MIRDcalc. (Bottom) OLINDA compared against MIRDcalc. Red indicates a dose coefficient estimate higher than that of MIRDcalc; blue indicates lower. Black indicates off-scale values ( > 300).
> 300).
All summary statistics (e.g., mean and SD) mentioned in the text consider the entire ICRP publication 128 radiopharmaceutical ensemble.
COMPARISON OF MIRDCALC AND IDAC-DOSE
Overall, the effective dose coefficients calculated from MIRDcalc and IDAC-Dose were in close agreement. On average, the IDAC-Dose effective dose estimates were marginally lower than MIRDcalc (mean  = −2.8, SD = 4.2; Eq. 13). The largest differences were observed for pulmonary perfusion imaging agents (e.g., 99mTc-macroaggregated albumin and 133Xe gas), for which the lung tissues were the critical organs; in these cases, the effective dose coefficients computed with IDAC-Dose were lower by approximately 5%–21%. These differences in effective dose reflected larger underlying differences in the aborbed doses computed for the bronchial and bronchiolar epithelial regions of the lung. Acute underestimates in the SAFs of ICRP publication 133 were originally published for the lung regions of the reference adult phantoms for low-energy electrons and photons (<100 keV), which would lead to underestimates of lung tissue–absorbed doses if uncorrected. These issues were corrected before generation of the MIRDcalc S-value database (22).
= −2.8, SD = 4.2; Eq. 13). The largest differences were observed for pulmonary perfusion imaging agents (e.g., 99mTc-macroaggregated albumin and 133Xe gas), for which the lung tissues were the critical organs; in these cases, the effective dose coefficients computed with IDAC-Dose were lower by approximately 5%–21%. These differences in effective dose reflected larger underlying differences in the aborbed doses computed for the bronchial and bronchiolar epithelial regions of the lung. Acute underestimates in the SAFs of ICRP publication 133 were originally published for the lung regions of the reference adult phantoms for low-energy electrons and photons (<100 keV), which would lead to underestimates of lung tissue–absorbed doses if uncorrected. These issues were corrected before generation of the MIRDcalc S-value database (22).
Since MIRDcalc and IDAC-Dose use the same phantoms and the same radionuclide decay data, very close agreement was expected. Exact agreement was not observed. Implementation details related to the blood source in each program are expected to be the major source of disagreement.
COMPARISON OF MIRDCALC AND ICRP PUBLICATION 128
Effective dose coefficients calculated from MIRDcalc differed considerably from those provided in ICRP publication 128 (adult subjects: mean  = +6.1, SD = 67), indicating a bias toward lower values than the ICRP publication 128 dose estimates. Absorbed dose coefficients for the urinary bladder wall were 2- to 3-fold higher for the ICRP publication 128 estimates than for MIRDcalc, for renally excreted radiopharmaceuticals (Figs. 2 and 3). This feature has been observed in many investigations and relates to conservative simplified methods for estimating the bladder wall irradiation from the bladder contents (18,19,23). ICRP publication 128 absorbed dose coefficients for the endosteal cells and adrenals showed consistent positive and negative biases, respectively, though to a lesser degree than seen with the urinary bladder. The definition of the radiosensitive endosteal cell target region has been refined in modern skeletal models and comprises a 50-μm layer at the surfaces of the trabecular spongiosa and cortical surfaces of the medullary cavities of the long bones of the limbs. This is a potential source of disagreement with dose coefficients derived in ICRP publication 128, which assume a 10-μm endosteal layer. The negatively biased adrenal-absorbed doses are likely due to the exaggeration of interorgan spacing in stylized phantoms. Cross-irradiation from the kidneys is an important contributor to the adrenal-absorbed dose for renally cleared agents and exacerbates the error due to adrenal–kidney spacing mismatch. Few other trends were evident between the absorbed dose coefficients of ICRP publication 128 and MIRDcalc. A similar variation was seen between the age-matched pediatric phantoms.
= +6.1, SD = 67), indicating a bias toward lower values than the ICRP publication 128 dose estimates. Absorbed dose coefficients for the urinary bladder wall were 2- to 3-fold higher for the ICRP publication 128 estimates than for MIRDcalc, for renally excreted radiopharmaceuticals (Figs. 2 and 3). This feature has been observed in many investigations and relates to conservative simplified methods for estimating the bladder wall irradiation from the bladder contents (18,19,23). ICRP publication 128 absorbed dose coefficients for the endosteal cells and adrenals showed consistent positive and negative biases, respectively, though to a lesser degree than seen with the urinary bladder. The definition of the radiosensitive endosteal cell target region has been refined in modern skeletal models and comprises a 50-μm layer at the surfaces of the trabecular spongiosa and cortical surfaces of the medullary cavities of the long bones of the limbs. This is a potential source of disagreement with dose coefficients derived in ICRP publication 128, which assume a 10-μm endosteal layer. The negatively biased adrenal-absorbed doses are likely due to the exaggeration of interorgan spacing in stylized phantoms. Cross-irradiation from the kidneys is an important contributor to the adrenal-absorbed dose for renally cleared agents and exacerbates the error due to adrenal–kidney spacing mismatch. Few other trends were evident between the absorbed dose coefficients of ICRP publication 128 and MIRDcalc. A similar variation was seen between the age-matched pediatric phantoms.
COMPARISON OF MIRDCALC AND OLINDA
Effective dose coefficients calculated with MIRDcalc also differed from those calculated with OLINDA (adult subjects: mean  = −11, SD = 50). The largest relative differences were generally observed for radiopharmaceuticals with gastrointestinal tract wall TIACs given by ICRP publication 128 (e.g., radioiodides)—which OLINDA was not designed to accommodate. OLINDA estimates for the urinary bladder–absorbed and bone endosteum–absorbed dose coefficients were positively biased relative to MIRDcalc. Compared with the stylized phantoms of ICRP publication 128, the phantoms used in OLINDA more closely match those used in MIRDcalc in terms of organ mass, contour, and spacing; however,
= −11, SD = 50). The largest relative differences were generally observed for radiopharmaceuticals with gastrointestinal tract wall TIACs given by ICRP publication 128 (e.g., radioiodides)—which OLINDA was not designed to accommodate. OLINDA estimates for the urinary bladder–absorbed and bone endosteum–absorbed dose coefficients were positively biased relative to MIRDcalc. Compared with the stylized phantoms of ICRP publication 128, the phantoms used in OLINDA more closely match those used in MIRDcalc in terms of organ mass, contour, and spacing; however,  was not significantly improved relative to
was not significantly improved relative to  .
.
COMPARISON OF RESULTS: WHAT CONSTITUTES REASONABLE AGREEMENT?
This investigation compares radiopharmaceutical dose coefficients derived from different reference phantoms with different software. Reference phantoms are designed to characterize the average individual of a specified age and sex—that is, the only criterion a phantom must meet to qualify as a reference phantom is that the organ masses defined within the phantom should match accepted reference values for the corresponding age and sex. Dose coefficients depend strongly on organ mass but are also influenced by other phantom-specific factors including organ shape, interorgan spacing, target volumes, degree of detail, and other factors, none of which are standardized. To generate phantom SAFs/S values needed for software using MIRD-style formalisms, Monte Carlo radiation transport simulations are performed within the phantoms. The details of the Monte Carlo simulations also lack standardization. Because such factors are not controlled, it is difficult to stipulate how well dose coefficients derived from different reference phantoms should agree.
It is generally accepted that variation in the body morphometry of specific patients—which may deviate substantially from the reference individual—imparts variation in organ-absorbed dose coefficients on the order of 20%–60% (8,24); this percentage range expressed in terms of Δ-values (Eq. 13) is approximately 18–47. We are unaware of any studies providing a ballpark guideline on agreement for reference phantoms, but the variation should be considerably lower because the organ masses are consistent (8). Therefore, we consider this range to be an upper limit regarding reasonable agreement. It is evident in Figures 2 and 3 that there are many instances in which  and
and  lie well outside this range, and many instances were also present for the radiopharmaceuticals not shown. To gain a more comprehensive but condensed view, we expanded the scope over all 116 cases examined and aggregated the Δ-values for each software comparison into histograms (Figs. 4 and 5, left panel). In each histogram, the red–green shading encompasses the Δ-values within the upper limit of the range of expected variation (Δ = 47). The left panel includes Δ-values for all available target regions that correspond directly between the software being compared.
lie well outside this range, and many instances were also present for the radiopharmaceuticals not shown. To gain a more comprehensive but condensed view, we expanded the scope over all 116 cases examined and aggregated the Δ-values for each software comparison into histograms (Figs. 4 and 5, left panel). In each histogram, the red–green shading encompasses the Δ-values within the upper limit of the range of expected variation (Δ = 47). The left panel includes Δ-values for all available target regions that correspond directly between the software being compared.
Distribution of log relative differences in organ-level absorbed dose coefficients for adult male phantoms. (Top) ICRP publication 128 compared against MIRDcalc. (Middle) IDAC-Dose compared against MIRDcalc. (Bottom) OLINDA compared against MIRDcalc. Red–green shaded region represents range of reasonable agreement discussed in text; percentage value overlying this region indicates fraction of Δ-values that fall within this range. Histogram bin width is 10.
Distribution of log relative differences in organ-level absorbed dose coefficients for adult female phantoms. (Top) ICRP publication 128 compared against MIRDcalc. Upper left histogram is negatively skewed because ICRP publication 128 dose estimates are derived from single 70-kg hermaphroditic adult phantom, whereas MIRDcalc, IDAC-Dose, and OLINDA use a 60-kg female phantom. (Middle) IDAC-Dose compared against MIRDcalc. (Bottom) OLINDA compared against MIRDcalc. Red-green shaded region represents range of reasonable agreement discussed in text; percentage value overlying this region indicates fraction of Δ-values that fall within this range. Histogram bin width is 10.
We questioned whether the outlying Δ-values (Figs. 4 and 5, left panel) were likely to be dosimetrically significant. For example, the critical organ (the organ receiving the highest absorbed dose) is important in planning administered activities for clinical trials for new imaging agents and is of greater importance if the agent is being used in a theranostic role (e.g., to plan therapeutic administrations based on pretherapy imaging). Is there consensus among the software and phantoms regarding absorbed dose to critical organs? To gain insight into this question, we restricted the histograms to include only the critical organ for each radiopharmaceutical (Figs. 4 and 5, middle panel). Notably, the histograms of  and
and  for the critical organs showed a reduced frequency of negative Δ-values. This finding indicates that OLINDA and ICRP publication 128, when compared with MIRDcalc, tend to provide higher estimates of the dose coefficients for critical organs. This tendency seems logical, because a large proportion of the radiopharmaceuticals included in ICRP publication 128 are rapidly excreted small molecules, which often irradiate the walled clearance organs to the largest extent, and because OLINDA and ICRP publication 128 are known to provide conservative overestimates for the walled organs (18,19,21,23). Analogous to the critical organ, we have also provided histograms that include only the organ contributing maximally to the effective dose (i.e., the maximal
for the critical organs showed a reduced frequency of negative Δ-values. This finding indicates that OLINDA and ICRP publication 128, when compared with MIRDcalc, tend to provide higher estimates of the dose coefficients for critical organs. This tendency seems logical, because a large proportion of the radiopharmaceuticals included in ICRP publication 128 are rapidly excreted small molecules, which often irradiate the walled clearance organs to the largest extent, and because OLINDA and ICRP publication 128 are known to provide conservative overestimates for the walled organs (18,19,21,23). Analogous to the critical organ, we have also provided histograms that include only the organ contributing maximally to the effective dose (i.e., the maximal  ; Figs. 4 and 5, right panel). When considering only critical organs or maximal contributors to the effective dose, agreement between MIRDcalc and IDAC-Dose improved such that no
; Figs. 4 and 5, right panel). When considering only critical organs or maximal contributors to the effective dose, agreement between MIRDcalc and IDAC-Dose improved such that no  lay outside the range of expected variation.
lay outside the range of expected variation.
LIMITATIONS AND FUTURE WORK
We provide revised reference dose estimates for the ICRP publication 110 phantoms (or other modernized phantoms) based on biokinetic data of ICRP publication 128. There are recognized uncertainties inherent in the ICRP 128 TIACs—uncertainties that exist largely because of the limited availability of quantitative biodistribution measurements for many radiopharmaceuticals. Aside from these uncertainties, the main limitation in our dosimetric evaluation strategy is that the TIACs were derived using older compartmental pharmacokinetic models or exponential retention functions intended for use with the Cristy–Eckerman stylized phantoms. Therefore, the dose coefficients provided here should not supersede those originally published in ICRP publication 128. Rather, the dose coefficients we present should be used for comparative purposes; for example, in software validation, as we show. A current effort within the ICRP focuses on updating the ICRP publication 128 biokinetic datasets to full compartmental models consistent with the updated phantoms. After favorable reception of these datasets by the field, the dose coefficients should be recalculated with MIRDcalc and other current dosimetry software.
The scope of this validation was limited to diagnostic cases only; future validation should investigate agreement of MIRDcalc with other software supporting personalized dosimetry for therapy and theranostic-use cases.
CONCLUSION
We report comprehensive testing and validation of MIRDcalc software based on comparison of dose coefficients for multiple radiopharmaceuticals across adult and pediatric phantoms, using validated data sources. Dose coefficients computed with MIRDcalc showed overall excellent agreement with other types of dosimetry software implementing the ICRP publication 110 series reference adult voxel phantoms and showed reasonable agreement with most dose coefficients derived using other reference phantoms.
DISCLOSURE
This research was funded in part through the NIH/NCI Cancer Center support grant P30 CA008748 and NIH U01 EB028234. Lukas Carter acknowledges support from the Ruth L. Kirschstein NRSA postdoctoral fellowship (NIH F32 EB025050). No other potential conflict of interest relevant to this article was reported.
ACKNOWLEGMENTS
This work was done in collaboration with the Society of Nuclear Medicine and Molecular Imaging MIRD Committee: Vikram Adhikarla, Rachel M. Barbee, Wesley E. Bolch, Yuni K. Dewaraja, William D. Erwin, Darrell R. Fisher, Roger W. Howell, Adam L. Kesner, Richard Laforest, Joseph G. Rajendran, George Sgouros, and Pat B. Zanzonico (chair). We thank Dr. Pat Zanzonico from Memorial Sloan Kettering Cancer Center for providing guidance and support to the MIRDcalc development effort.
Footnotes
Published online Jun. 2, 2023.
- © 2023 by the Society of Nuclear Medicine and Molecular Imaging.
REFERENCES
- Received for publication April 4, 2022.
- Revision received March 21, 2023.












