Abstract
Purpose
Delineation of tumour boundaries is important for quantification of [18F]fluoro-2-deoxy-D-glucose (FDG) positron emission tomography (PET) studies and for definition of biological target volumes in radiotherapy. Several (semi-)automatic tumour delineation methods have been proposed, but these methods differ substantially in estimating tumour volume and their performance may be affected by imaging parameters. The main purpose of this study was to explore the performance dependence of various (semi-)automatic tumour delineation methods on different imaging parameters, i.e. reconstruction parameters, noise levels and tumour characteristics, and thereby the need for standardization or inter-institute calibration.
Methods
Six different types of delineation methods were evaluated by assessing accuracy and precision in estimating tumour volume from simulations and phantom experiments. The evaluated conditions were various tumour sizes, iterative reconstruction algorithm settings and image filtering, tumour to background ratios (TBR), noise levels and region growing initializations.
Results
The accuracy of all automatic delineation methods was influenced when imaging parameters were varied. The performance of all tumour delineation methods depends on variation of TBR, image resolution and image noise level, and to a lesser extent on number of iterations during image reconstruction or the initialization method of the region generation. For sphere sizes larger than 20 mm diameter a contrast-oriented method provided the most accurate results, on average, over all simulated conditions. For threshold-based methods the accuracy of tumour delineation improved after image denoising/filtering.
Conclusion
The accuracy and precision of all studied tumour delineation methods was affected by physiological and imaging parameters. The latter illustrates the need for optimizing imaging parameters and/or for careful calibration and optimization of delineation methods.
Similar content being viewed by others
Introduction
Quantitative analysis of oncological positron emission tomography (PET) studies, e.g. for response assessment, target definition for radiotherapy and glycolytic tumour volume measurements, usually involves delineation of tumour boundaries and several tumour segmentation methods have been reported [1–3]. Accurate and reproducible delineation of tumours may enhance accuracy and precision of quantitative PET studies required for response monitoring purposes. In addition, there is growing interest in the use of PET for tumour delineation in radiotherapy [4, 5]. In this case, accurate tumour delineation is extremely important in order to focus the dose on viable tumour tissue, thereby sparing surrounding normal tissue and allowing a higher radiation dose to the target region.
[18F]Fluoro-2-deoxy-D-glucose (FDG) is most widely used for detection, staging and treatment monitoring as well as for determining boundaries of the target volume in radiotherapy [6–9]. [18F]FDG tumour delineation methods range from visual (manual) approaches to (semi-)automatic algorithms [1–3, 10]. These studies have compared tumour contours obtained with [18F]FDG PET, CT, MRI and/or pathologic data, reporting substantially different tumour volumes. Furthermore, manual delineation methods strongly depend on the experience of the physician and on the specific contouring protocol used [11]. This may lead to high variation in [18F]FDG-based gross tumour volume (GTV) delineation. There is general consensus that (semi-)automatic delineation methods may reduce this variability.
Numerous factors could affect accuracy of PET quantification, i.e. scanner or image resolution, scanner type and settings of image reconstruction algorithms, image filtering, level of image noise, and tumour characteristics [12]. To date, however, evaluation of the various factors affecting performance of tumour delineation or volume of interest (VOI) methods is still limited. Consequently, VOI methods are often being used without proper validation or optimization for the specific scanner at a particular site [13]. Therefore, the primary objective of this study was to explore the performance of various commonly used (semi-)automatic VOI methods depending on imaging parameters, i.e. image reconstruction settings and filtering, image noise levels, and tumour characteristics. The secondary objective is thus to demonstrate the potentially large errors that may occur when using these methods in a non-calibrated or non-standardized manner and to illustrate the need for inter-institute calibration when using PET for the assessment of ‘metabolic’ tumour volumes.
Materials and methods
Simulation experiments
The same procedures for performing simulations as described in [14] were used. The method is based on a computer-generated image of the thorax that was derived from a summed 2-D dynamic [18F]FDG scan of a typical patient (i.e. all image data collected from 15 to 60 min post-injection are summed into a single image). Next, spheres of various sizes (10, 20, 30, 40 and 50 mm diameter), simulating tumours, were placed in both lung and mediastinum, applying tumour to (lung) background ratios (TBR) of 4, 8 and 12. These images were then forward-projected using a rotation-based forward projector to generate sinograms. Poisson noise was then applied to these sinograms, such that reconstructed images showed voxel variances of ~22 and ~47% coefficients of variation (COV) in homogeneous background areas (liver or mediastinum). The transmission scan obtained during the same clinical patient study was used to incorporate the effects of photon attenuation. These computer-generated sinograms were reconstructed using attenuation-weighted ordered subsets expectation maximization (OSEM) with 16 subsets and 1, 2, 4 or 6 iterations. An image matrix size of 256 × 256 was used, corresponding to a voxel size of 2.56 × 2.56 × 2.56 mm3. For each combination of tumour size, resolution, TBR, noise level and number of iterations, 100 reconstructed images were generated from 100 ‘noisy’ realizations (i.e. Poisson noise was applied 100 times) of a sinogram to evaluate both accuracy and precision of each volumetric tumour delineation method. Reconstructed images were used without and with additional smoothing using a 3-D isotropic Gaussian kernel of 5 and 7 mm full-width at half-maximum (FWHM), thereby reducing both resolution and image noise. Finally, all images (either with or without the Gaussian filtering) were evaluated with and without use of an edge-preserving bilateral filter for further denoising in order to reduce statistical noise in the images. This filter consists of a Gaussian smoothing filter (10 mm FWHM) which is multiplied by a second Gaussian function that penalizes the filter based on differences in voxel intensity or standardized uptake value (SUV) between neighbouring voxels (sigma: 1.5).The algorithm is implemented using two iterations, exactly as described in [3].
Phantom experiments
To further evaluate the various volume delineation methods, a National Electrical Manufacturers Association (NEMA) image quality phantom was used, containing six spheres with volumes ranging from 0.53 to 26.52 ml (having diameters of 10, 13, 17, 22, 28 and 37 mm, respectively). The background compartment was filled with an [18F]FDG solution of 5 kBq/ml and spheres were filled with an [18F]FDG solution of 25 or 45 kBq/ml (simulating a TBR of approximately 5 or 9, respectively).
The phantom was first scanned using an ECAT EXACT HR+ scanner (Siemens/CTI, Knoxville, TN, USA) [15]. The PET study started with a 10-min transmission scan, after which an emission scan was acquired in 3-D mode. This scan consisted of 20 frames, each with 300 kilocounts, corresponding to the average number of counts observed in 3 min of a typical oncological [18F]FDG study. Data were reconstructed using OSEM with 2 iterations and 16 subsets. Additional smoothing was applied using a 5 mm FWHM Gaussian filter. Resulting images consisted of an image matrix size of 256 × 256 × 63 and a voxel size of 2.57 × 2.57 × 2.43 mm3.
The same phantom was also scanned on a GEMINI TF PET/CT system (Philips Healthcare, Highland Heights, Ohio, USA) [16], again with 20 frames and the same counts (300 kilocounts) for all frames. Data were reconstructed using a time-of-flight (TOF) reconstruction algorithm (blob-based OSEM TOF) using standard reconstruction settings, as these cannot easily be modified by the user. Resulting images consisted of an image matrix size of 144 × 144 × 44 and a voxel size of 4.00 × 4.00 × 4.00 mm3.
After reconstruction, the same edge-preserving bilateral filter as applied in the simulations was used in order to reduce noise in the images from both the HR+ and GEMINI TF phantom experiments.
Data analysis
Six different types of (semi-)automatic VOI methods were used to determine tumour volumes in the PET images:
-
1.
Fixed threshold range of 41–70% of maximum voxel value within tumour (VOI41, VOI50, VOI70). This method applies a threshold based on a percentage (i.e. here 41–70%) of the maximum voxel intensity within the tumour [14]. Next this threshold is used to delineate the tumour.
-
2.
Adaptive threshold range of 41–70% of maximum voxel value within tumour (VOIA41, VOIA50, VOIA70). This method is similar to the fixed threshold method, except that it adapts the threshold relative to the local average background, thereby correcting for the contrast between tumour and local background [14]. For example, the A50 contour value corresponds to a value at 50% of the sum of the maximum voxel value and the local background value. The latter value is derived from ‘background’ voxels that are identified as those voxels located on a single voxel thick shell at 2.5 cm from the edge of a 70% of maximum pixel value isocontour, excluding all voxels with an SUV larger than 2.5.
-
3.
Contrast-oriented method (VOISchaefer). This algorithm [1] uses a correction by measuring the mean of 70% SUVmax and background activity for various sphere sizes. Regression coefficients are calculated, which represent the relationship between optimal threshold and image contrast for various sphere sizes. This threshold equation is given by:
$$ Threshol{d_{{optimal}}} = A \times meanSU{V_{{70\% }}} + B \times Background $$where A and B were fitted using phantom studies [1]. In general, different values are applied for sphere diameters smaller and larger than 3 cm diameter. In the present paper, we calibrated this method, i.e. we determined the A and B values that are specific for each of the simulated image resolutions and for both PET systems included in this study.
-
4.
Background-subtracted relative threshold level (RTL) method (VOIRTL). This method is an iterative method based on a convolution of the point spread function (PSF) that takes into account the differences between various sphere sizes and the scanner resolution [2]. The optimal RTL depends on diameter size, but not on TBR. For the phantom experiments we used the PSF measured on each scanner and for the simulations we used the simulated image resolutions.
-
5.
Gradient-based watershed segmentation method (GradWT). This method uses two steps before calculating the VOI. First, this method calculates a gradient image on which one ‘seed’ is placed in the tumour and another one in the background. Next, a watershed (WT) algorithm [3] is used to grow the seeds in the gradient basins, thereby creating boundaries on the gradient edges. In our presentation, the watershed continues to grow the gradient basins until all voxels are either classified as ‘tumour’ or ‘non-tumour (background)’. The voxel will be assigned to tumour in case two watersheds are competing for the same voxel, i.e. ‘border’ voxels are assigned as tumour.
-
6.
Absolute SUV (SUV2.5). Normalized (SUV) voxel intensities at a chosen absolute threshold are used to delineate tumour. An SUV of 2.5 was used as it might properly differentiate between benign and malignant lesions and might also be used for GTV delineation [17]. However, this threshold of 2.5 remains arbitrary.
For all methods the maximum intensity (i.e. maximum voxel value) within a sphere was identified. In addition, the maximum voxel value was obtained by applying a cross-shaped pattern or VOI that could be less sensitive to noise. This method searches for the region with the (local) average maximum intensity, based on the average of seven neighbouring voxels, which was then used as maximum or ‘peak’ value.
All delineation methods were evaluated by assessing accuracy (bias) and precision (standard deviation, SD) of estimating the volume. Per cent bias was defined as \( (\frac{{Volum{e_{{measured}}}}}{{Volum{e_{{true}}}}} - 1) \times 100\% \). Note that bias may be negative, indicating underestimation of the sphere volume. Accuracy and precision of the estimated volumes were calculated using averages and SD over 100 simulations or over 20 phantom images. In addition, for the simulations, the mean of absolute differences between percentage bias of the 30-mm sphere and percentage bias of other sphere sizes (i.e. 20, 40 and 50 mm) was reported to assess the variation in bias across the different tumour sizes.
Results
Simulation experiment: general results
Tumour boundaries from automatic delineation methods were affected by the choice of various settings, i.e. sphere size, TBR, reconstruction settings, image resolution and noise levels. Figure 1 shows bias as function of tumour size for simulation data in both lung (Fig. 1a) and mediastinum (Fig. 1b). Data presented in Fig. 1 were generated using a TBR of 8 and simulated sinograms were reconstructed using OSEM with 4 iterations, 16 subsets and 5 mm FWHM Gaussian post-smoothing. Most methods showed an underestimation of tumour volume for both regions (i.e. approximately −5 to −25% for the largest sphere), with the exception of GradWT that showed an overestimation (>23% for the 20- to 50-mm diameter spheres in both regions). In the case of the smallest sphere, none of the methods could define tumour volume accurately, especially in the mediastinum (with biases up to 45% for VOIRTL). VOISchaefer, however, was able to accurately derive tumour volume in most cases within about 10%. Only the five best performing methods (i.e. GradWT, VOI41, VOIA41, VOIRTL and VOISchaefer), i.e. absolute bias <15% for spheres ≥20 mm in the lung, were evaluated further for various image characteristics.
In general, both HR+ (Fig. 2a, b) and GEMINI TF (Fig. 2c, d) phantom studies showed similar trends as in simulations, indicating that simulations are suitable for evaluating the impact of image characteristics on performance of VOI methods. As simulations are more flexible in generating different data sets with multiple noisy replicates [14], the effects of varying imaging parameters and image characteristics were assessed using simulation data only.
Accuracy of delineation methods as function of sphere size for phantom data with various TBR; (a) and (b) represent data for HR+; (c) and (d) represent data for GEMINI TF; (a) and (c) represent data for TBR of 5; (b) and (d) represent data for TBR of 9. Note that some data points fall outside the scale of the figure (> ±31%) and that some data points overlap
Simulation experiment: effects of reconstruction settings
There were only minor differences when image reconstruction settings were varied. In general, each delineation method showed small differences in bias (<3% for 20- to 50-mm diameter spheres) when varying the numbers of iterations.
Simulation experiment: effects of TBR
The accuracy of tumour delineation methods depends on TBR as indicated in Table 1 (Supplementary Fig. 1). For most methods (GradWT, VOI41, VOIA41, VOIRTL and VOISchaefer) bias increased when TBR decreased with the largest differences in bias between TBR values of 4 and 8. Only GradWT showed a large increase in the mean of absolute difference in bias amongst various tumour sizes when TBR decreased from 8 to 4 (4.6%, other methods: <0.6%). Bias improved by only ~3% when TBR was increased from 8 to 12.
Simulation experiment: effects of spatial resolution
Large differences were observed between smoothed and non-smoothed images for all methods (Table 1 and Supplementary Fig. 2). Smoothing the images improved the bias in delineated tumour volumes as obtained with VOI41, VOIA41, VOIRTL, GradWT and VOISchaefer to at most about 10% for all, except the smallest sphere sizes. For GradWT the mean of absolute difference amongst various tumour sizes improved when smoothing was applied. Large differences were observed between non-smoothed computer-generated images of high noise levels and smoothed images for all VOI methods.
Simulation experiment: effects of noise level, image filtering and region growing initialization
Slightly better accuracy for percentage threshold methods was observed as well as a slight improvement in SD of about 1% when using a cross-shaped pattern instead of the maximum voxel value (Table 1). Bias obtained with VOI41 and VOIA41 improved with approximately 5% for each sphere size when compared to using a single voxel maximum value.
Use of the bilateral filter improved bias by approximately 10% for VOI41, VOIA41 and VOIRTL. The resulting measured volume was close to the actual volume for spheres ≥20 mm (bias <5%). In contrast, GradWT did not show any effect of the edge-preserving bilateral filter for larger sphere sizes (30–50 mm).
For VOI41, VOIA41, VOIRTL and VOISchaefer bias increased for higher noise levels. The negative bias obtained with these four VOI methods was increased with 15% compared to normal noise levels (Table 1 and Supplementary Fig. 3a). Effects of applying the edge-preserving bilateral filter to data simulated at higher noise levels showed similar improvements as were observed for data simulated at normal noise levels (Table 1 and Supplementary Fig. 3b).
Phantom experiment
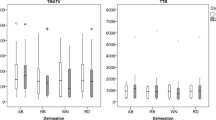
Figure 2 summarizes the results of the phantom experiment simulating effect of different TBRs (5 and 9) and scanner resolutions. Performance of GradWT and VOI41 improved by increasing resolution and TBR. VOIA41, VOIRTL and VOISchaefer were only slightly affected by increased TBR and resolutions. VOI41 showed poor performance for small sphere sizes, especially for a TBR of 5. Precision of the delineation methods was slightly different for the two scanners (data not shown). For example for the HR+, the largest sphere (37-mm diameter) showed an SD of 1.0–1.7% for TBR of 9, whereas the small sphere (13-mm diameter) showed an SD of 9.0–13%. For the GEMINI TF PET/CT system similar SDs were found (1.3–2.1% and 5.5–11%, respectively).
Discussion
In this study, the main objective was to explore the performance dependence of several (semi-)automatic delineation methods [1–3, 14, 17] as function of different image characteristics in the case of [18F]FDG scans. For all methods substantial variation in bias was observed, but the different methods showed different sensitivities to variations in sphere size, TBR, reconstruction settings, image resolution and noise levels. Secondly, the paper intends to examine the potentially large errors that may occur when using these methods in a non-standardized or non-calibrated method. We also explored VOISchaefer without calibration and observed very high bias in measured volume (i.e. >38% bias for a 30-mm diameter sphere in the lung), which was strongly reduced after calibration (<7%). Therefore, in the present paper, in line with the recommendations pointed out in [1, 18], only VOISchaefer with calibration was used. An alternative approach would be to harmonize the image quality (i.e. spatial resolution, TBR and quantitative accuracy) across various sites as attempted by the recently published European Association of Nuclear Medicine (EANM) guidelines [19]. This approach would only be required when using methods that cannot be calibrated for specific imaging parameters, e.g. threshold-based methods, either with or without background corrections, in order to ensure inter-institute comparability of PET-based tumour volume assessments.
All delineation methods could not define tumour volume accurately for all sphere sizes, i.e. SUV2.5 showed large bias in estimating tumour volume (i.e. >25% bias for a 30-mm diameter sphere in the lung). As there are no ‘normal’ values of SUV that can be applied to every situation, it has been shown previously [5] that SUV2.5 can often fail to produce accurate tumour volumes, e.g. when the physiological background activity lies above the fixed threshold. The remaining methods (GradWT, VOI41, VOIA41, VOIRTL and VOISchaefer) provided acceptable accuracy, i.e. for spheres >20 mm they showed biases smaller than 18 and 23% for lung and mediastinum, respectively.
Fixed threshold-based methods (i.e. 41–70% of maximum voxel value) strongly depended on the threshold level chosen. Delineated volumes for higher thresholds are obviously smaller, resulting in underestimation of volumes. Advanced adaptive threshold-based methods (e.g. VOISchaefer) do not use a fixed threshold level, but also correct for background activity, and tumour volume or mean tumour intensity. The presented results showed minor dependence on noise, spatial resolution, acquisition parameters and reconstruction settings for VOISchaefer, as was expected when calibrating the method. Overall VOISchaefer seems to perform well over various simulated imaging characteristics.
Simulation studies
Based on the initial results, only five methods (GradWT, VOI41, VOIA41, VOIRTL and VOISchaefer) were evaluated further in relation to various imaging parameters. The accuracy of these methods was affected by tumour size, TBR, image resolution and noise level. By optimizing the imaging parameters the accuracy of the delineated volume estimates increased for all VOI methods investigated.
There was a large difference in accuracy of delineated volume between unsmoothed and smoothed images and/or at various noise levels. All VOI methods tested showed a poor performance for non-smoothed data, which is likely caused by the high noise levels in the computer-generated images. There are several possible causes for the noise dependence of various VOI methods. First of all, methods which use a percentage of maximum uptake to define the final contour are likely to be more sensitive to noise as noise may result in an upward bias of the maximum value. Consequently, the upward bias in the maximum value may result in higher isocontour values and thus in smaller volumes. Secondly, noise will impact the accuracy and precision of any 3-D region growing technique. Therefore, noise will directly impact the granularity of the observed contours and thereby accuracy of observed VOI. When noise levels become too high 3-D region growing algorithms may fail to generate a meaningful VOI. However, the difference in accuracy of delineated volume between smoothed (additional 5 mm FWHM) and more smoothed (additional 7 mm FWHM) was much less (Table 1 and Supplementary Fig. 2). In general, good accuracy (bias <12%) for the delineation methods was found when using 7 mm FWHM smoothed images. However, smoothing with 7 mm FWHM could induce partial volume effects and loss of detail [14]. The latter effect also explains why most methods have difficulty in providing accurate tumour volumes for small spheres. A lower resolution will also degrade the gradient between tumour and non-tumour tissue and, consequently, it will be more difficult for any VOI method to delineate the tumour boundaries. In the presence of lower gradients small uncertainties in the actual threshold being used by the VOI method for tumour delineation (as is the case for most VOI methods used in this study) could result in larger ‘displacements’ of the generated contour. In the case of gradient-based methods it is obvious that lower gradients will result in less accurate assessments of the position of the steepest gradient and thus in increased uncertainty and reduced accuracy of this method at lower resolutions.
The results obtained by changing noise levels and degree of smoothing indicate that there is a sensitive trade-off between noise and resolution. Ideally, images should have high spatial resolution and very low noise levels. However, in clinical practice some filtering is applied to reduce noise levels. As explained above, elevated noise levels may also hamper (semi-)automated tumour delineation and, especially when expected tumour sizes are large and have high FDG uptake, some filtering may be helpful to generate reliable tumour volume estimates. Yet, filtering degrades image resolution which in turn hampers tumour delineation for smaller tumours (e.g. <15 mm diameter) with lower uptakes (TBR <4). Therefore, in practice the trade-off between noise and resolution should be carefully considered and optimization of imaging parameters in combination with calibrating the VOI method (when possible for the envisioned method) is needed depending on the scanner, tracer, VOI method and tumour type and location.
Effects of an edge-preserving bilateral filter for denoising images were also investigated (Table 1 and Supplementary Fig. 3b). After applying the filter to data sets at two noise levels, the accuracy of all methods, except for GradWT, improved. Again this may illustrate the sensitivity of most VOI methods to noise. The lack of improvement of GradWT is not fully clear, but a possible explanation for overestimation of tumour volume could be that in our implementation a voxel will be assigned to tumour in case two watersheds are competing for the same voxel, i.e. border voxels are assigned as tumour. Further work is ongoing to enhance the performance of this method, e.g. by allowing for fractional voxels and/or using a higher image matrix size (upsampling). In addition, in this paper we explored the effects of noise reduction using Gaussian and bilateral filtering. It should be noted that both these filters do not take the Poisson nature of noise into account, i.e. the variance is proportional to the underlying signal. Possibly, tumour delineations will benefit from more sophisticated filtering approaches that include an estimate of local variance.
When using an iterative reconstruction algorithm, both quantitative accuracy and noise level depend on number of iterations. A higher number of iterations not only improves convergence and image contrast, but also increases image noise. Only small differences in bias (<3% lower) were observed when varying the number of iterations for each VOI method. This indicates that the chosen reconstruction setting does not show a large effect on accuracy of measured tumour volumes. Similar results were shown in a previous study [20] that more extensively evaluated the effects of various reconstruction algorithms and settings. It was shown that accuracy of measured volume varies only slightly with image reconstruction algorithm and smaller spheres (i.e. <2 ml) were affected more than larger spheres. The latter was also seen in the present study, i.e. accuracy of tumour volume was better for larger (>30 mm) than for smaller spheres.
Using the cross-shaped pattern to identify an averaged maximum or peak value and its location provided similar results as those based on the maximum (single) voxel value. Accuracy of GradWT and VOIRTL methods was similar for all spheres compared to using a single voxel maximum value. This can easily be understood as both methods do not use the maximum (or peak) voxel value. On the other hand, as can be expected, VOI41 and VOIA41 showed a small improvement by 3–6% (Table 1). In addition, the SD of these methods improved slightly when using the cross-shaped pattern, probably because the effects of noisy voxels are reduced by using an average value. Using a cross-shaped pattern did improve performance of percentage threshold-based methods and therefore it is recommended to use this approach for initialization, especially when percentage threshold-based methods are used.
Phantom studies
Similar to what was observed in the simulation studies there was a limitation in defining volumes for the smaller spheres (diameter <15 mm, Fig. 2). Therefore, the smallest sphere gave large biases for all methods (sometimes >70%). For all delineation methods, the best performance was observed for sphere sizes larger than 15 mm diameter. For the HR+ and the GEMINI TF, VOISchaefer seemed to be the best method on average.
Moreover, this study showed that effective threshold-based methods that correct for local background activity (i.e. VOIA41 and VOIRTL), contrast-oriented (i.e. VOISchaefer) as well as gradient-based methods are useful for defining tumour volume. However, optimal percentage threshold level and/or optimal settings strongly depend on imaging parameters. Likewise, VOISchaefer needs reassessment of the method’s parameters as function of image characteristics (mainly image resolution). This implies that calibration of VOI methods and/or (in combination with) optimization of PET procedures is required when PET images are used for tumour delineation [19].
Limitations
Firstly, tumours in both experiments were represented by homogeneous 3-D spheres, thereby excluding effects of tumour shape and heterogeneity. Therefore, even methods that showed good performance in the present paper should be used with care and need to be supervised in the case of (non-spherical) tumours showing heterogeneous tracer uptake. Widely available methods that can accurately deal with variation in imaging characteristics and tracer uptake heterogeneity are needed. In this respect the fuzzy locally adaptive Bayesian method published by Hatt et al. [21] appears to be very promising. Secondly, in the phantom experiments, but not in the simulations, background activity was uniform around the tumour. This is usually not the case in actual human PET studies and higher local uptake (e.g. due to inflammation) may result in errors when defining tumour contours. On the other hand, for the phantom experiments the wall of the spheres, resulting in a shell of ‘zero’ activity around the spheres, may have affected performance evaluation [22]. Yet, phantom study results were similar to those seen in simulation results and vice versa. Finally, this study focused on tumours located in the thorax. Therefore, all methods should be evaluated further for other body regions and using clinical data. Even with these ‘simple’ conditions, however, it is clear that differences in image characteristics, caused by differences in reconstruction settings, image filtering and noise levels, can have a pronounced effect on performance of the (semi-)automatic delineation methods investigated, although magnitude and direction of those effects may be different among (semi-)automatic delineation methods.
Conclusion
Differences in imaging parameters can have a pronounced effect on the performance of (semi-)automated methods to delineate tumours. This implies that PET data collection and image reconstruction procedures and the (semi-)automated tumour delineation methods need to be standardized and calibrated for each scanner for reproducible and accurate tumour delineations.
References
Schaefer A, Kremp S, Hellwig D, Rübe C, Kirsch CM, Nestle U. A contrast-oriented algorithm for FDG-PET-based delineation of tumour volumes for the radiotherapy of lung cancer: derivation from phantom measurements and validation in patient data. Eur J Nucl Med Mol Imaging 2008;35:1989–99.
van Dalen JA, Hoffmann AL, Dicken V, Vogel WV, Wiering B, Ruers TJ, et al. A novel iterative method for lesion delineation and volumetric quantification with FDG PET. Nucl Med Commun 2007;28:485–93.
Geets X, Lee JA, Bol A, Lonneux M, Grégoire V. A gradient-based method for segmenting FDG-PET images: methodology and validation. Eur J Nucl Med Mol Imaging 2007;34:1427–38.
Zaidi H, El Naqa I. PET-guided delineation of radiation therapy treatment volumes: a survey of image segmentation techniques. Eur J Nucl Med Mol Imaging 2010;37:2165–87.
Nestle U, Weber W, Hentschel M, Grosu AL. Biological imaging in radiation therapy: role of positron emission tomography. Phys Med Biol 2009;54:R1–R25.
Fletcher JW, Djulbegovic B, Soares HP, Siegel BA, Lowe VJ, Lyman GH, et al. Recommendations on the use of 18F-FDG PET in oncology. J Nucl Med 2008;49:480–508.
de Geus-Oei LF, van der Heijden HF, Corstens FH, Oyen WJ. Predictive and prognostic value of FDG-PET in nonsmall-cell lung cancer: a systematic review. Cancer 2007;110:1654–64.
Erdi YE. The use of PET for radiotherapy. Curr Med Imaging Rev 2007;3:3–16.
Juweid ME, Cheson BD. Positron-emission tomography and assessment of cancer therapy. N Engl J Med 2006;354:496–507.
Roels S, Slagmolen P, Nuyts J, Lee JA, Loeckx D, Maes F, et al. Biological image-guided radiotherapy in rectal cancer: is there a role for FMISO or FLT, next to FDG? Acta Oncol 2008;47:1237–48.
MacManus M, Nestle U, Rosenzweig KE, Carrio I, Messa C, Belohlavek O, et al. Use of PET and PET/CT for radiation therapy planning: IAEA expert report 2006–2007. Radiother Oncol 2009;91:85–94.
Boellaard R. Standards for PET image acquisition and quantitative data analysis. J Nucl Med 2009;50 Suppl 1:11S–20S.
Hatt M, Visvikis D, Le Rest CC. Autocontouring versus manual contouring. J Nucl Med 2011;52:658–9.
Boellaard R, Krak NC, Hoekstra OS, Lammertsma AA. Effects of noise, image resolution, and ROI definition on the accuracy of standard uptake values: a simulation study. J Nucl Med 2004;45:1519–27.
Brix G, Zaers J, Adam LE, Bellemann ME, Ostertag H, Trojan H, et al. Performance evaluation of a whole-body PET scanner using the NEMA protocol. National Electrical Manufacturers Association. J Nucl Med 1997;38:1614–23.
Surti S, Kuhn A, Werner ME, Perkins AE, Kolthammer J, Karp JS. Performance of Philips Gemini TF PET/CT scanner with special consideration for its time-of-flight imaging capabilities. J Nucl Med 2007;48:471–80.
Paulino AC, Koshy M, Howell R, Schuster D, Davis LW. Comparison of CT- and FDG-PET-defined gross tumor volume in intensity-modulated radiotherapy for head-and-neck cancer. Int J Radiat Oncol Biol Phys 2005;61:1385–92.
Nestle U, Kremp S, Schaefer-Schuler A, Sebastian-Welsch C, Hellwig D, Rübe C, et al. Comparison of different methods for delineation of 18F-FDG PET-positive tissue for target volume definition in radiotherapy of patients with non-small cell lung cancer. J Nucl Med 2005;46:1342–8.
Boellaard R, O’Doherty MJ, Weber WA, Mottaghy FM, Lonsdale MN, Stroobants SG, et al. FDG PET and PET/CT: EANM procedure guidelines for tumour PET imaging: version 1.0. Eur J Nucl Med Mol Imaging 2010;37:181–200.
Daisne JF, Sibomana M, Bol A, Doumont T, Lonneux M, Grégoire V. Tri-dimensional automatic segmentation of PET volumes based on measured source-to-background ratios: influence of reconstruction algorithms. Radiother Oncol 2003;69:247–50.
Hatt M, Cheze Le Rest C, Albarghach N, Pradier O, Visvikis D. PET functional volume delineation: a robustness and repeatability study. Eur J Nucl Med Mol Imaging 2011;38:663–72.
Hofheinz F, Dittrich S, Pötzsch C, Hoff J. Effects of cold sphere walls in PET phantom measurements on the volume reproducing threshold. Phys Med Biol 2010;55:1099–113.
Acknowledgements
The authors would like to thank N.J. Hoetjes for assistance in acquiring phantom data and the radiochemistry staff for production of isotopes. This study was performed within the framework of CTMM, the Center for Translational Molecular Medicine, AIRFORCE project (grant 03O-103). Patsuree Cheebsumon was supported by a scholarship from the National Science and Technology Development Agency of the Royal Thai Government.
Conflicts of interest
None.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Fig. 1
Accuracy of selected delineation methods as function of sphere size for a TBR of 4 (a) and 12 (b). Note that some data fall outside the scale of the figure (> ±34%) and that some symbols overlap. (JPEG 15 kb)
Supplementary Fig. 2
Accuracy of selected delineation methods as function of sphere size for a non-smoothed data, b smoothed data with 5 mm FWHM Gaussian filter and c smoothed data with 7 mm FWHM Gaussian filter. Note that some data fall outside the scale of the figure (> ±50%) and that some symbols overlap. (JPEG 22 kb)
Supplementary Fig. 3
Accuracy of selected delineation methods as function of sphere size for the higher noise level data obtained by adding ~47% COV of noise (a) and with additional image denoising using an edge-preserving bilateral filter (b). Note that some data fall outside the scale of the figure (> ±30%) and that some symbols overlap. (JPEG 15 kb)
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Cheebsumon, P., Yaqub, M., van Velden, F.H.P. et al. Impact of [18F]FDG PET imaging parameters on automatic tumour delineation: need for improved tumour delineation methodology. Eur J Nucl Med Mol Imaging 38, 2136–2144 (2011). https://doi.org/10.1007/s00259-011-1899-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00259-011-1899-5